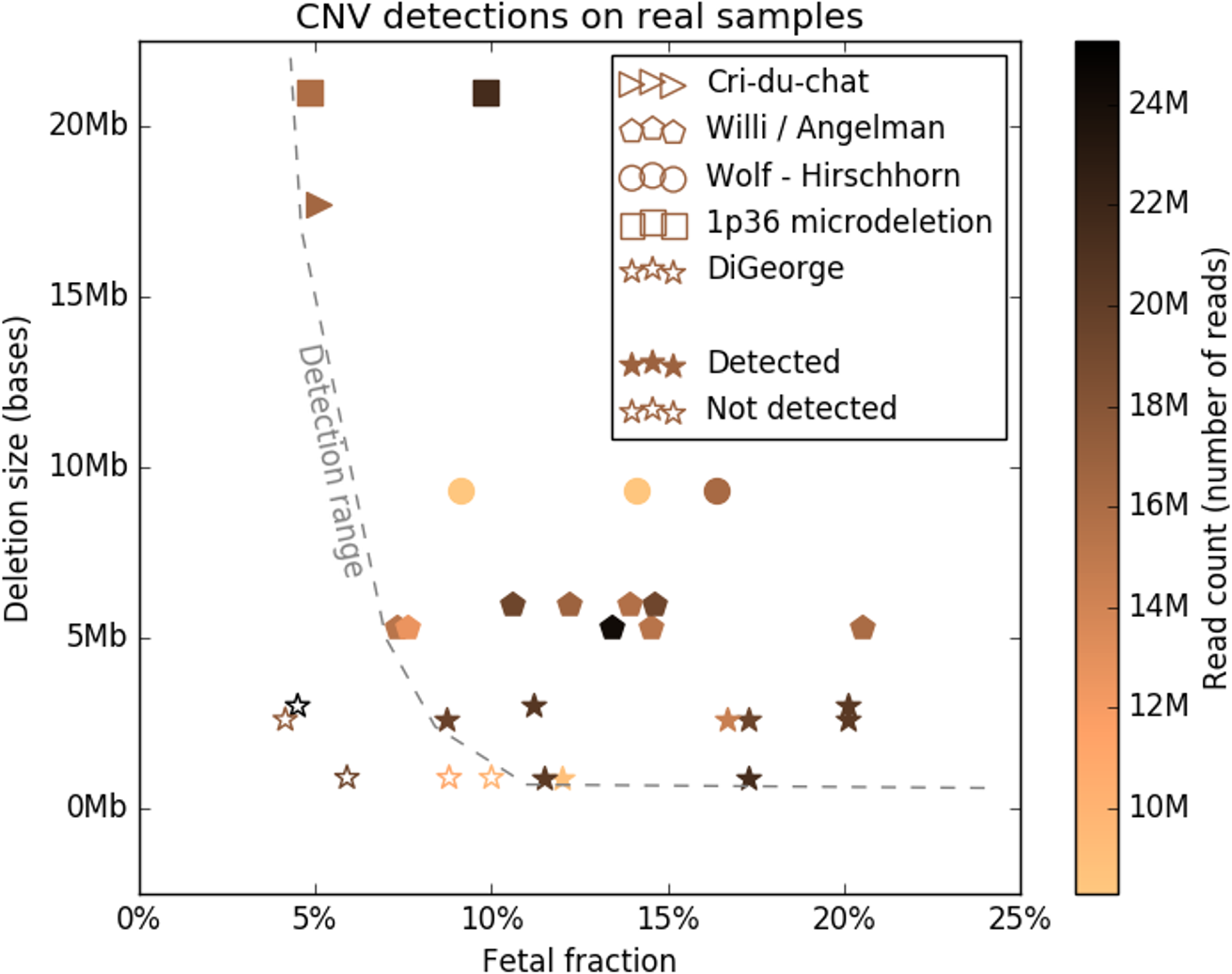
To study the detection limits of chromosomal microaberrations in non-invasive prenatal testing with aim for five target microdeletion syndromes, including DiGeorge, Prader-Willi/Angelman, 1p36, Cri-Du-Chat, and Wolf-Hirschhorn syndromes. We used known cases of pathogenic deletions from ISCA database to specifically define regions critical for the target syndromes. Our approach to detect microdeletions, from whole genome sequencing data, is based on sample normalization and read counting for individual bins.
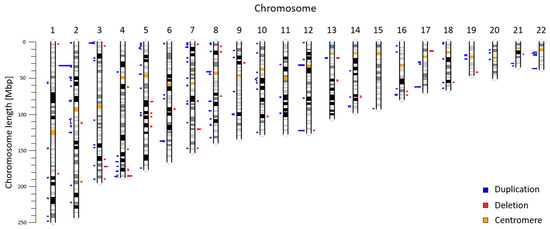
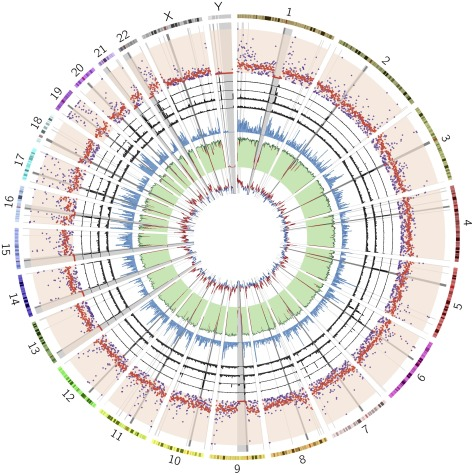
Read MoreWe reanalyzed NIPT genomic data from 5018 patients to evaluate CNV aberrations in the Slovak population. Our analysis of autosomal chromosomes identified 225 maternal CNVs (47 deletions; 178 duplications) ranging from 600 to 7820 kbp. In summary, we identified 129 likely benign variants, 13 variants of uncertain significance, and 83 likely pathogenic variants. In this study, we use NIPT as a valuable source of population specific data. Our results suggest the utility of genomic data from commercial CNV analysis test as background for a population study.
Read MoreLow-coverage massively parallel genome sequencing for non-invasive prenatal testing (NIPT) of common aneuploidies is one of the most rapidly adopted and relatively low-cost DNA tests. Since aggregation of reads from a large number of samples allows overcoming the problems of extremely low coverage of individual samples, we describe the possible re-use of the data generated during NIPT testing for genome scale population specific frequency determination of small DNA variants, requiring no additional costs except of those for the NIPT test itself.
Read More