Non-invasive prenatal testing (NIPT) by low coverage genomic sequencing: Detection limits of screened chromosomal microdeletions
Marcel Kucharik1,2, Andrej Gnip3,4, Michaela Hyblova3,4, Jaroslav Budis1,2,5, Lucia Strieskova1, Maria Harsanyova1,6, Ondrej Pös1,6, Zuzana Kubiritova1,6,7, Jan Radvanszky1,7, Gabriel Minarik3,4, Tomas Szemes1,2,6
- 1Geneton Ltd., Bratislava, Slovakia
- 2Comenius University Science Park, Bratislava, Slovakia
- 3Medirex Inc., Bratislava, Slovakia.
- 4TrisomyTest Ltd., Bratislava, Slovakia
- 5Slovak Centre of Scientific and Technical Information, Bratislava, Slovakia
- 6Department of Molecular Biology, Faculty of Natural Sciences, Comenius University, Bratislava, Slovakia
- 7Institute of Clinical and Translational Research, Biomedical Research Center, Slovak Academy of Sciences, Bratislava, Slovakia
Abstract
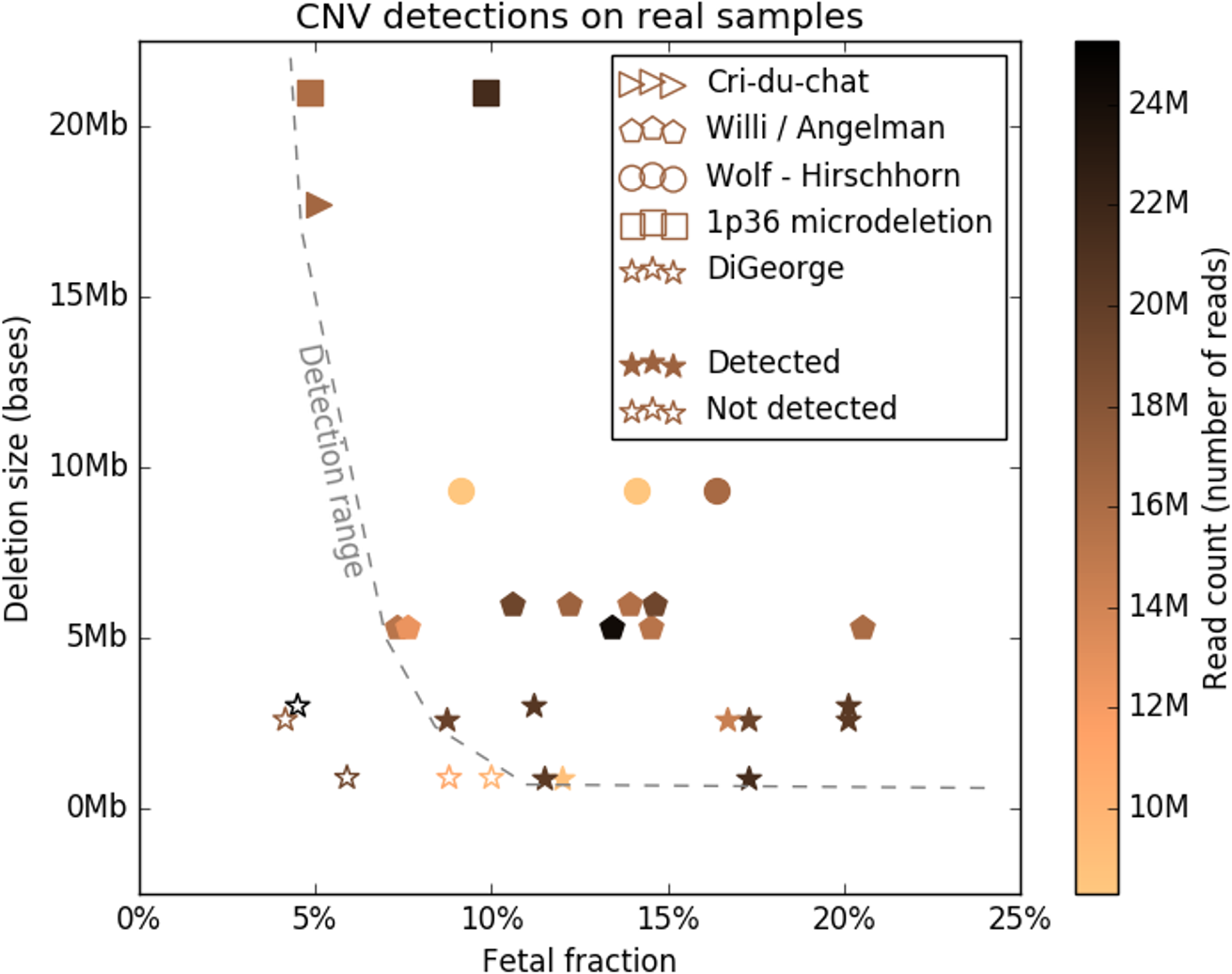
To study the detection limits of chromosomal microaberrations in non-invasive prenatal testing with aim for five target microdeletion syndromes, including DiGeorge, Prader-Willi/Angelman, 1p36, Cri-Du-Chat, and Wolf-Hirschhorn syndromes. We used known cases of pathogenic deletions from ISCA database to specifically define regions critical for the target syndromes. Our approach to detect microdeletions, from whole genome sequencing data, is based on sample normalization and read counting for individual bins. We performed both an in-silico study using artificially created data sets and a laboratory test on mixed DNA samples, with known microdeletions, to assess the sensitivity of prediction for varying fetal fractions, deletion lengths, and sequencing read counts. The in-silico study showed sensitivity of 79.3% for 10% fetal fraction with 20M read count, which further increased to 98.4% if we searched only for deletions longer than 3Mb. The test on laboratory-prepared mixed samples was in agreement with in-silico results, while we were able to correctly detect 24 out of 29 control samples. Our results suggest that it is possible to incorporate microaberration detection into basic NIPT as part of the offered screening/diagnostics procedure, however, accuracy and reliability depends on several specific factors.