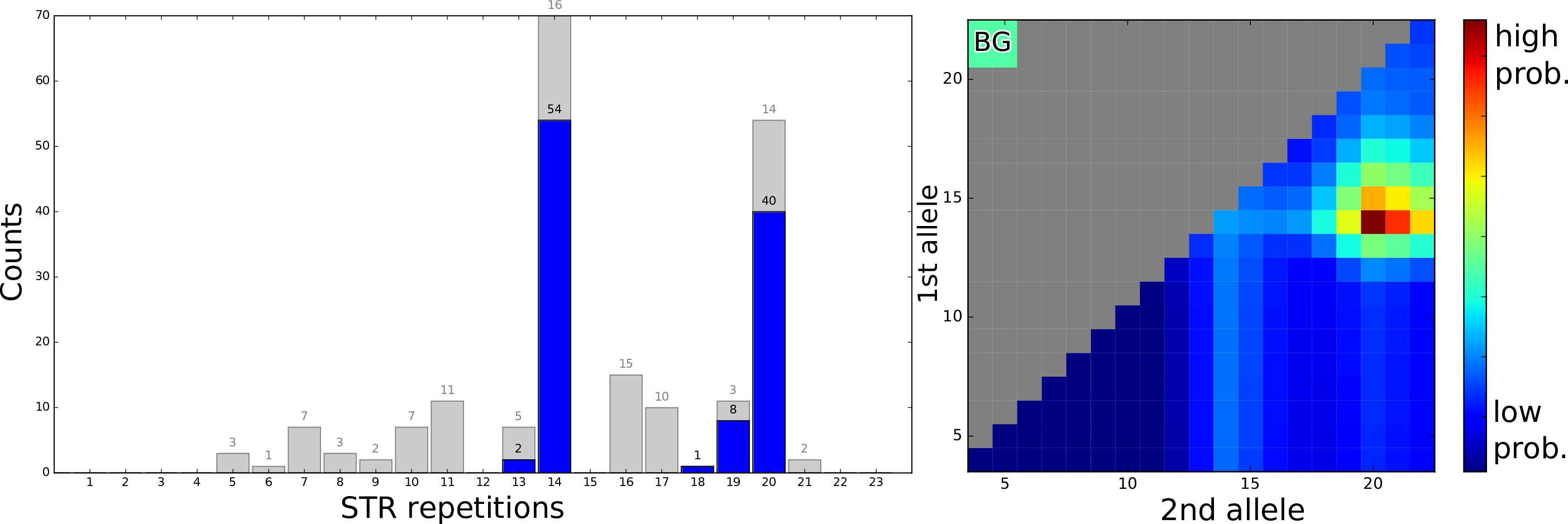
Short tandem repeats (STRs) are stretches of repetitive DNA in which short sequences, typically made of 2-6 nucleotides, are repeated several times. STRs are highly relevant in molecular-genetic applications – several STRs are linked to various genetic disorders.
Dante is an alignment-free algorithm for genotyping and characterizing STR alleles based on sequence reads originating from STR loci of interest. This method accounts for natural deviations from the expected sequence, such as variation in the repeat count, sequencing errors, ambiguous bases, stutter effect, complex loci containing several different motifs, even STR expansions that, according to the conventional view, cannot be fully captured by inherently short massive parallel sequencing reads.
In all our tests, Dante outperformed state-of-the-art genotyping tools, HipSTR (Li, 2009) and GATK (McKenna, 2010). Furthermore, Dante was able to predict allele expansions in all tested clinical cases.
Dante generates user friendly report with specific characteristics of the genotyped alleles together with the information about the expansions. All this combined makes Dante specifically suitable for evaluation of clinically relevant STR loci in molecular diagnostic applications.
More info in publication: https://doi.org/10.1093/bioinformatics/bty791
Dante can be downloaded from GitHub: https://github.com/jbudis/dante